Introduction
This tutorial aims to describe the different features of the R
package TDLM. The main purpose of the TDLM
package is to provide a rigorous framework for fairly comparing trip
distribution laws and models (Lenormand et
al., 2016). This general framework is based on a two-step
approach for generating mobility flows by separating the trip
distribution law, such as gravity or intervening opportunities, from the
modeling approach used to generate flows from this law.
A short note on terminology
This framework is part of the four-step travel model. It corresponds to the second step, called trip distribution, which aims to match trip origins with trip destinations. The model used to generate trips or flows, and more generally the degree of interaction between locations, is often referred to as a spatial interaction model. Depending on the research area, a matrix or a network formalism may be used to describe these spatial interactions. Origin-Destination matrices (or trip tables) are common in geography or transportation, while in statistical physics or complex systems studies, the term mobility networks is usually preferred.
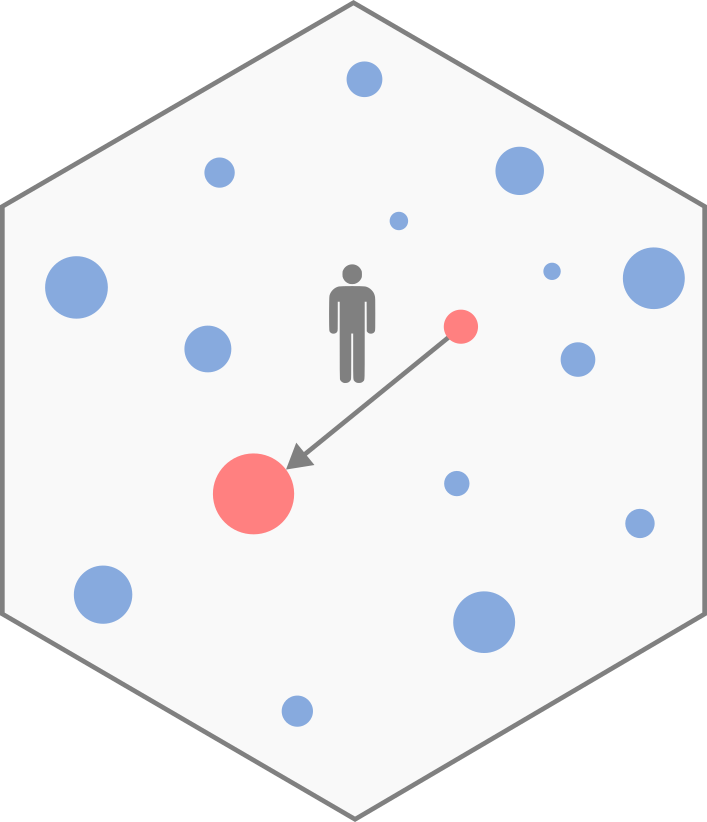
Origin–Destination matrix
The description of movements within a given area is represented by an Origin-Destination (OD) matrix. The area of interest is divided into locations, and represents the volume of flows between location and location . This volume typically corresponds to the number of trips or a commuting flow (i.e., the number of individuals living in and working in ). The OD matrix is square, contains only positive values, and may have a zero diagonal (Figure 1).
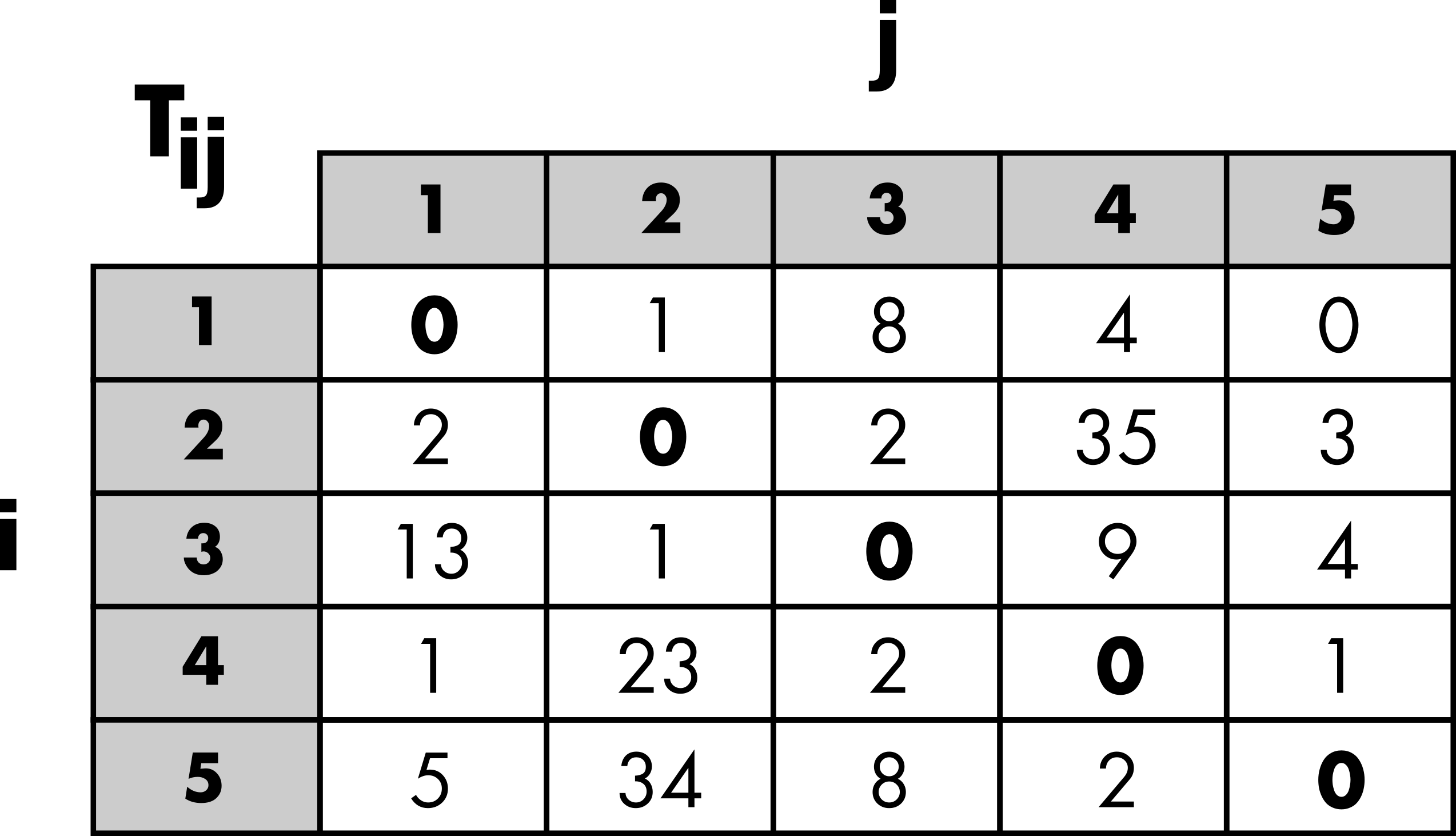
Aggregated inputs information
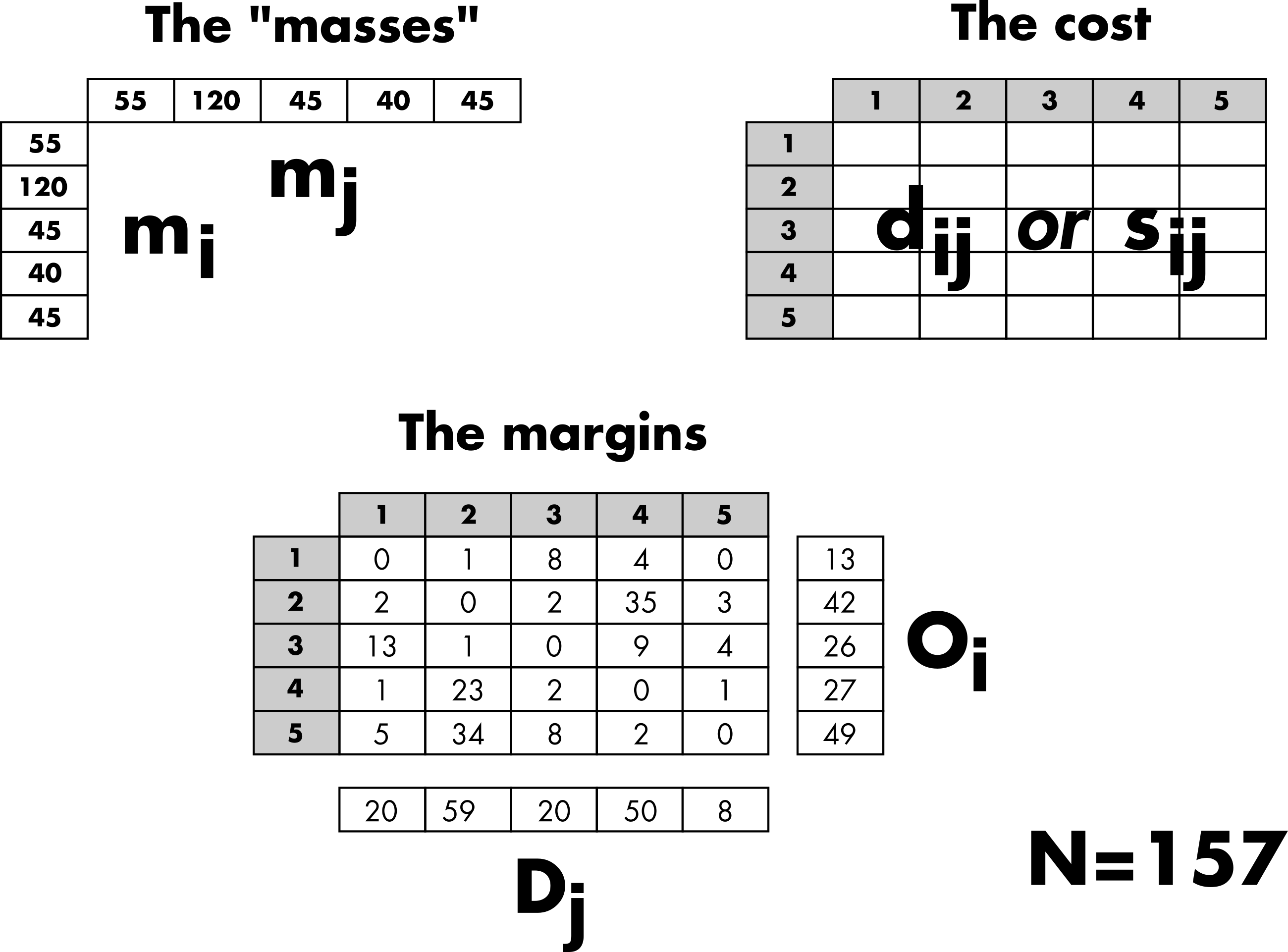
Three categories of inputs are typically considered to simulate an OD matrix (Figure 2). The masses and distances are the primary ingredients used to generate a matrix of probabilities based on a given distribution law. Thus, the probability of observing a trip from location to location depends on the masses, the demand at the origin (), and the offer at the destination (). Typically, population is used as a surrogate for demand and offer. The probability of movement also depends on the costs, which are based on the distance between locations or the number of opportunities between locations, depending on the chosen law (more details are provided in the next “Trip distribution laws” section). In general, the effect of the cost can be adjusted with a parameter.
The margins are used to generate an OD matrix from the matrix of probabilities while preserving the total number of trips (), the number of outgoing trips (), and/or the number of incoming trips () (more details are provided in the “Constrained distribution models” section).
Trip distribution laws
The purpose of a trip distribution law is to estimate the probability that, out of all the possible travels in the system, there is one between location and location . This probability is asymmetric in and , as are the flows themselves. It takes the form of a square matrix of probabilities. This probability is normalized across all possible pairs of origins and destinations, such that . Therefore, a matrix of probabilities can be obtained by normalizing any OD matrix (Figure 3).

As mentioned in the previous section, most trip distribution laws depend on the demand at the origin (), the offer at the destination (), and a cost to move from to . There are two major approaches for estimating the probability matrix. The traditional gravity approach, in analogy with Newton’s law of gravitation, is based on the assumption that the amount of trips between two locations is related to their populations and decays according to a function of the distance between locations. In contrast to the gravity law, the laws of intervening opportunities hinge on the assumption that the number of opportunities between locations plays a more important role than the distance (Lenormand et al., 2016). This fundamental difference between the two schools of thought is illustrated in Figure 4.
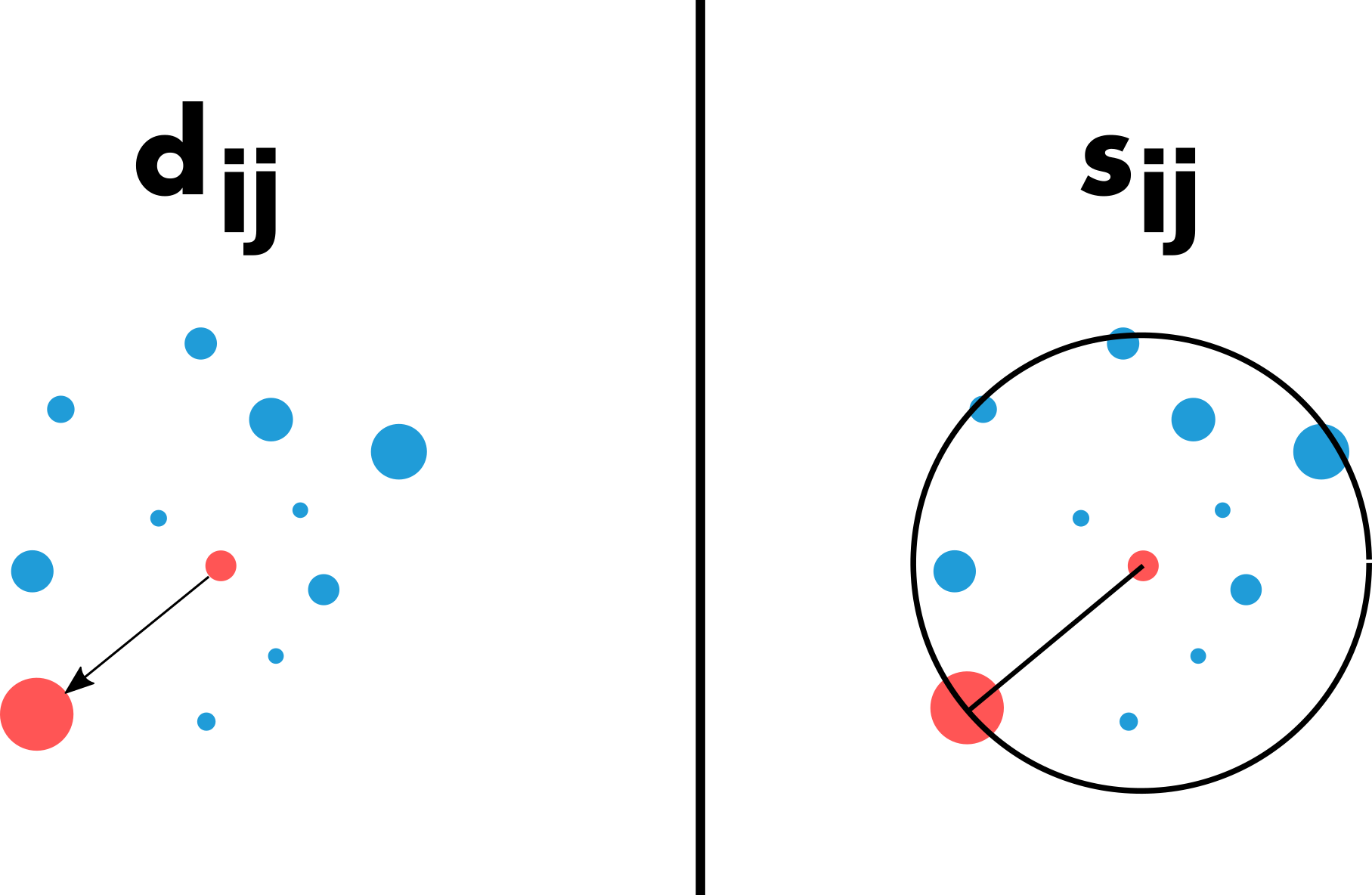
It is important to note that the effect of the cost between locations (distance or number of opportunities) can usually be adjusted with a parameter, which can be calibrated automatically or by comparing the simulated matrix with observed data.
Constrained distribution models
The purpose of the trip distribution models is to generate an OD matrix by drawing at random trips from the trip distribution law respecting different level of constraints according to the model. We considered four different types of models in this package. As can be observed in Figure 5, the four models respect different level of constraints from the total number of trips to the total number of out-going and in-coming trips by locations (i.e. the margins).
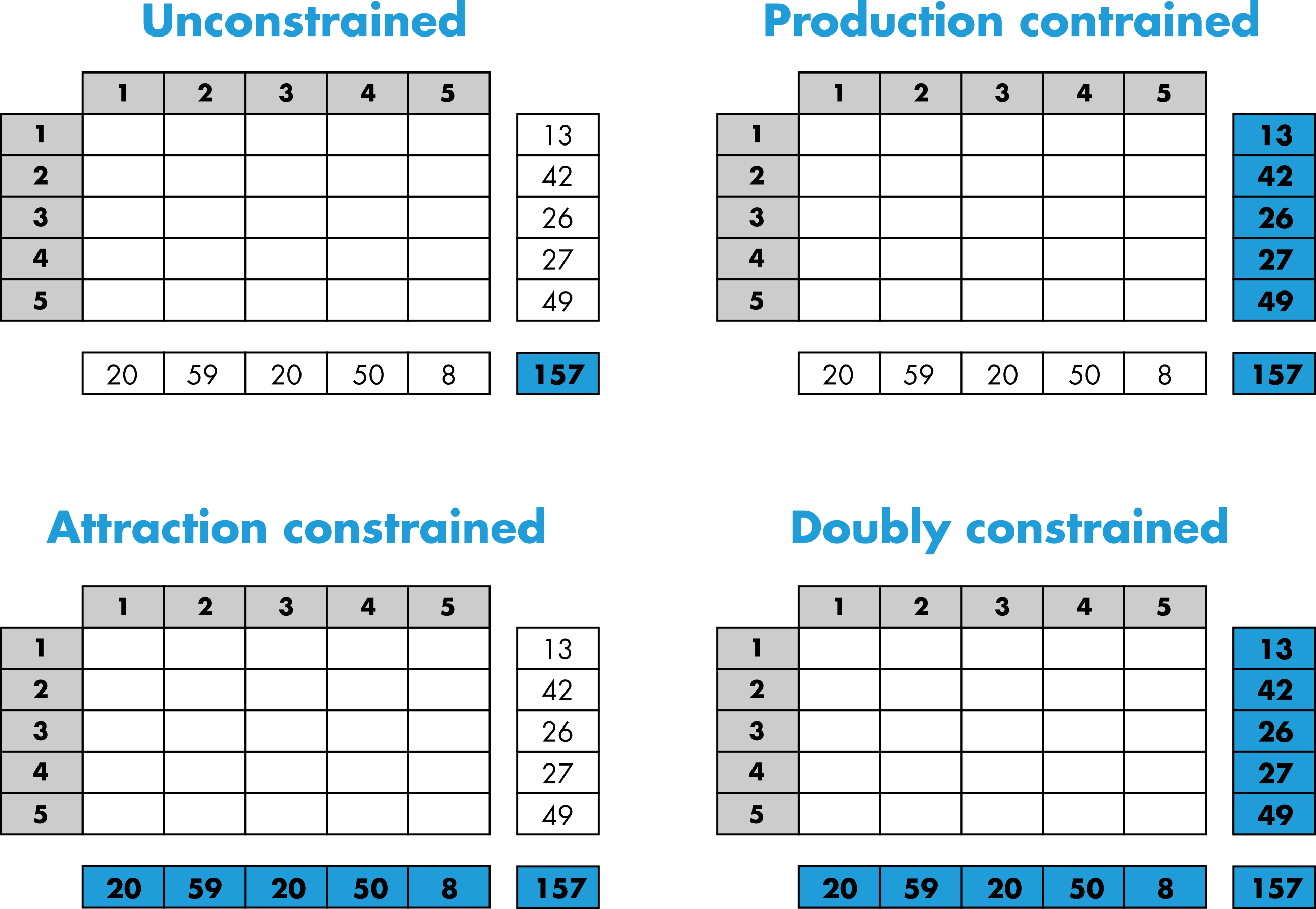
More specifically, the volume of flows is generated from the matrix of probability with multinomial random draws that will take different forms according to the model used (Lenormand et al., 2016). Therefore, since the process is stochastic, each simulated matrix is unique and composed of integers. Note that it is also possible to generate an average matrix from the multinomial trials.
Goodness-of-fit measures
Finally, the trip distribution laws can be calibrated, and both the trip distribution laws and models can be evaluated by comparing a simulated matrix with the observed matrix . These comparisons rely on various goodness-of-fit measures, which may or may not account for the distance between locations. These measures are described above.
Example of commuting in Kansas
Data
In this example, we will use commuting data from US Kansas in 2000 to
illustrate
the main functions of the package. The dataset comprises three tables
providing information on commuting flows between the 105 US Kansas
counties in 2000. The observed OD matrix od is a
zero-diagonal square matrix of integers. Each element of the matrix
represents the number of commuters between a pair of US Kansas
counties.
data(od)
od[1:5, 1:5]## 20001 20003 20005 20007 20009
## 20001 0 71 0 0 0
## 20003 236 0 0 0 0
## 20005 0 0 0 0 0
## 20007 0 0 0 0 8
## 20009 0 0 0 11 0
dim(od)## [1] 105 105The aggregated data are composed of the distance matrix,
data(distance)
distance[1:5, 1:5]## 20001 20003 20005 20007 20009
## 20001 0.00000 36.50943 182.9291 306.8503 308.8995
## 20003 36.50943 0.00000 146.4335 317.5593 303.2348
## 20005 182.92913 146.43350 0.0000 389.5330 319.5319
## 20007 306.85034 317.55926 389.5330 0.0000 139.0661
## 20009 308.89947 303.23478 319.5319 139.0661 0.0000
dim(distance)## [1] 105 105and the masses and margins contained in the data.frame mass.
data(mass)
mass[1:10,]## Population Out-commuters In-commuters
## 20001 14385 1267 1343
## 20003 8110 1346 361
## 20005 16774 1065 1247
## 20007 5307 260 201
## 20009 28205 1129 1324
## 20011 15379 662 761
## 20013 10724 1148 984
## 20015 59482 14182 3579
## 20017 3030 681 93
## 20019 4359 486 180
dim(mass)## [1] 105 3
mi <- as.numeric(mass[,1])
names(mi) <- rownames(mass)
mj <- mi
Oi <- as.numeric(mass[,2])
names(Oi) <- rownames(mass)
Dj <- as.numeric(mass[,3])
names(Dj) <- rownames(mass)The data must always be based on the same number of locations sorted
in the same
order. The function check_format_names
can be used to verify the validity of all inputs before running the main
functions of the package.
check_format_names(vectors = list(mi = mi, mj = mj, Oi = Oi, Dj = Dj),
matrices = list(od = od, distance = distance),
check = "format_and_names")## The inputs passed the format_and_names checks successfully!Optional spatial information are also provided here. county is a spatial object containing the geometry of the 105 US Kansas counties in 2000.
## Simple feature collection with 10 features and 4 fields
## Geometry type: MULTIPOLYGON
## Dimension: XY
## Bounding box: xmin: -99.0334 ymin: 36.99796 xmax: -94.6139 ymax: 40.0006
## Geodetic CRS: WGS 84
## ID Longitude Latitude Area geometry
## 1016 20001 -95.30137 37.88581 1307.667 MULTIPOLYGON (((-95.08805 3...
## 983 20003 -95.29334 38.21429 1512.337 MULTIPOLYGON (((-95.07771 3...
## 869 20005 -95.31288 39.53194 1125.682 MULTIPOLYGON (((-95.56751 3...
## 1064 20007 -98.68482 37.22888 2941.524 MULTIPOLYGON (((-98.52686 3...
## 962 20009 -98.75650 38.47904 2330.541 MULTIPOLYGON (((-99.03239 3...
## 1017 20011 -94.84928 37.85522 1653.609 MULTIPOLYGON (((-94.61413 3...
## 843 20013 -95.56416 39.82657 1480.469 MULTIPOLYGON (((-95.77332 3...
## 1011 20015 -96.83911 37.78125 3744.168 MULTIPOLYGON (((-96.52571 3...
## 974 20017 -96.59395 38.30205 2013.697 MULTIPOLYGON (((-96.81972 3...
## 1079 20019 -96.24535 37.15007 1669.418 MULTIPOLYGON (((-96.52495 3...
plot(county)
coords and coords_xy are two dataframes providing longitude/latitude and X/Y coordinates for each location, respectively.
coords[1:10,]## Longitude Latitude
## 20001 -95.30137 37.88581
## 20003 -95.29334 38.21429
## 20005 -95.31288 39.53194
## 20007 -98.68482 37.22888
## 20009 -98.75650 38.47904
## 20011 -94.84928 37.85522
## 20013 -95.56416 39.82657
## 20015 -96.83911 37.78125
## 20017 -96.59395 38.30205
## 20019 -96.24535 37.15007
coords_xy[1:10,]## X Y
## 20001 -10608899 4563329
## 20003 -10608005 4609773
## 20005 -10610183 4798171
## 20007 -10985547 4471108
## 20009 -10993524 4647367
## 20011 -10558574 4559025
## 20013 -10638154 4840804
## 20015 -10780080 4548659
## 20017 -10752789 4622229
## 20019 -10713984 4460066Extract Additional Spatial Information
The functions extract_distances,
extract_opportunities,
and extract_spatial_information
can be used to extract matrices of distances and the number of
intervening opportunities.
The first function computes distances in kilometers between pairs of locations based on geographical coordinates. It can calculate either great-circle distances, using longitude/latitude coordinates and the Haversine formula
haversine_d <- extract_distances(coords = coords,
method = "Haversine")
haversine_d[1:5, 1:5]## [,1] [,2] [,3] [,4] [,5]
## [1,] 0.00000 36.50943 182.9291 306.8503 308.8995
## [2,] 36.50943 0.00000 146.4335 317.5593 303.2348
## [3,] 182.92913 146.43350 0.0000 389.5330 319.5319
## [4,] 306.85034 317.55926 389.5330 0.0000 139.0661
## [5,] 308.89947 303.23478 319.5319 139.0661 0.0000
distance[1:5, 1:5]## 20001 20003 20005 20007 20009
## 20001 0.00000 36.50943 182.9291 306.8503 308.8995
## 20003 36.50943 0.00000 146.4335 317.5593 303.2348
## 20005 182.92913 146.43350 0.0000 389.5330 319.5319
## 20007 306.85034 317.55926 389.5330 0.0000 139.0661
## 20009 308.89947 303.23478 319.5319 139.0661 0.0000or Euclidean distances based on X/Y coordinates
xy_d <- extract_distances(coords = coords_xy,
method = "Euclidean")
oldmar <- par()$mar
par(mar = c(4.5, 6, 1, 1))
plot(haversine_d, xy_d, xlim=c(0,900), ylim=c(0,900),
type="p", pch=16, cex=2, lty=1, lwd=3,
col="steelblue3", axes=FALSE, xlab="", ylab="")
axis(1, cex.axis=1.2)
axis(2, cex.axis=1.2, las=1)
mtext("Haversine (km)", 1, line = 3.25, cex = 1.75)
mtext("Euclidean (km)", 2, line = 4, cex = 1.75)
box(lwd=1.5)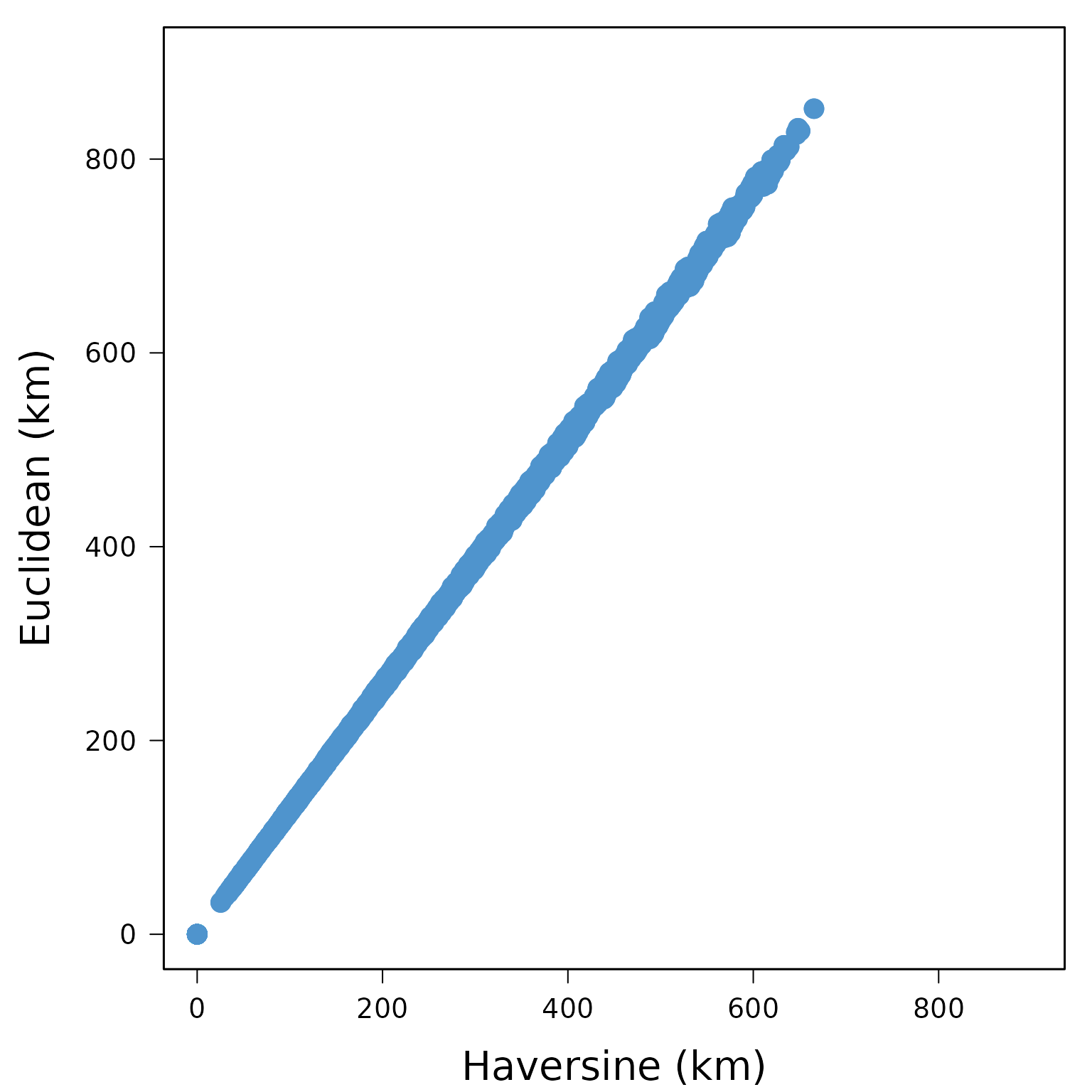
par(mar = oldmar)The second function computes the number of opportunities between pairs of locations. For a given pair of locations, the number of opportunities between the origin and the destination is based on the number of opportunities within a circle of radius equal to the distance between the origin and the destination, centered at the origin. The number of opportunities at the origin and the destination are not included. In our case, the number of inhabitants () is used as a proxy for the number of opportunities.
sij <- extract_opportunities(opportunity = mi,
distance = distance,
check_names = TRUE)
sij[1:5, 1:5]## 20001 20003 20005 20007 20009
## 20001 0 16997 1445725 2358187 2363494
## 20003 0 0 1374401 2403828 2354815
## 20005 1311224 1240896 0 2454129 2346302
## 20007 1433163 1481954 2489482 0 634786
## 20009 1778443 1651945 1820549 344665 0The last function takes as input a spatial object containing the
geometry of the locations that can be handled by the sf package. It returns
a matrix of great-circle distances between locations (expressed in km).
An optional id
can also be provided to be used as names for the outputs.
spi <- extract_spatial_information(county, id = "ID")
sp_d <- spi$distance
sp_d[1:5, 1:5]## 20001 20003 20005 20007 20009
## 20001 0.0000000000 0.0003285808 0.001646171 0.003446635 0.003505694
## 20003 0.0003285808 0.0000000000 0.001317793 0.003531738 0.003473271
## 20005 0.0016461705 0.0013177925 0.000000000 0.004083384 0.003600990
## 20007 0.0034466353 0.0035317381 0.004083384 0.000000000 0.001252210
## 20009 0.0035056940 0.0034732710 0.003600990 0.001252210 0.000000000
distance[1:5, 1:5]## 20001 20003 20005 20007 20009
## 20001 0.00000 36.50943 182.9291 306.8503 308.8995
## 20003 36.50943 0.00000 146.4335 317.5593 303.2348
## 20005 182.92913 146.43350 0.0000 389.5330 319.5319
## 20007 306.85034 317.55926 389.5330 0.0000 139.0661
## 20009 308.89947 303.23478 319.5319 139.0661 0.0000This function also allows extracting the surface area of each
location (in square kilometers), which can be useful to calibrate the
trip distribution
laws’ parameter value (see below).
mean(spi$surface)## [1] 2028.05Run functions
The main function of the package is run_law_model. The function has two sets of arguments, one for the law and another one for the model. The inputs (described above) necessary to run this function depends on the law (either the matrix of distances or number of opportunities can be used, or neither of them for the uniform law) and on the model and its associated constraints (number of trips, out-going trips and/or in-coming trips). The example below will generate three simulated ODs with the normalized gravity law with an exponential distance decay function (Lenormand et al., 2016) and the Doubly Constrained Model.
res <- run_law_model(law = "NGravExp",
mass_origin = mi,
mass_destination = mj,
distance = distance,
opportunity = NULL,
param = 0.01,
write_proba = TRUE,
model = "DCM",
nb_trips = NULL,
out_trips = Oi,
in_trips = Dj,
average = FALSE,
nbrep = 3)The output is an object of class TDLM. In this case it
is a list of matrices composed of the three simulated matrices
(replication_1, replication_2 and
replication_3), the matrix of probabilities (called
proba) associated with the law and returned only if
write_proba = TRUE. The objects of class TDLM
contain a table info summarizing the simulation run.
print(res)## Argument Value
## 1 Law NGravExp
## 2 Model DCM
## 3 #Replications 3
## 4 #Parameters 1
## 5 Parameter 0.01
str(res)## List of 5
## $ info :'data.frame': 5 obs. of 2 variables:
## ..$ Argument: chr [1:5] "Law" "Model" "#Replications" "#Parameters" ...
## ..$ Value : chr [1:5] "NGravExp" "DCM" "3" "1" ...
## $ replication_1: num [1:105, 1:105] 0 23 4 0 2 16 5 103 5 6 ...
## ..- attr(*, "dimnames")=List of 2
## $ replication_2: num [1:105, 1:105] 0 24 5 0 2 15 4 104 6 6 ...
## ..- attr(*, "dimnames")=List of 2
## $ replication_3: num [1:105, 1:105] 0 24 4 0 2 16 5 105 5 6 ...
## ..- attr(*, "dimnames")=List of 2
## $ proba : num [1:105, 1:105] 0.00 4.31e-05 2.16e-05 3.90e-06 1.81e-05 ...
## ..- attr(*, "dimnames")=List of 2
## - attr(*, "class")= chr [1:2] "TDLM" "list"
## - attr(*, "from")= chr "run_law_model"
## - attr(*, "proba")= logi TRUEThis simulation run was based on one parameter value. It is possible
to use a vector instead of a scalar for the param
argument.
res <- run_law_model(law = "NGravExp",
mass_origin = mi,
mass_destination = mj,
distance = distance,
opportunity = NULL,
param = c(0.01,0.02),
write_proba = TRUE,
model = "DCM",
nb_trips = NULL,
out_trips = Oi,
in_trips = Dj,
average = FALSE,
nbrep = 3)In this case a list of list of matrices will be returned (one for each parameter value).
print(res)## Argument Value
## 1 Law NGravExp
## 2 Model DCM
## 3 #Replications 3
## 4 #Parameters 2
## 5 Parameter 1 0.01
## 6 Parameter 2 0.02
str(res)## List of 3
## $ info :'data.frame': 6 obs. of 2 variables:
## ..$ Argument: chr [1:6] "Law" "Model" "#Replications" "#Parameters" ...
## ..$ Value : chr [1:6] "NGravExp" "DCM" "3" "2" ...
## $ parameter_1:List of 4
## ..$ replication_1: num [1:105, 1:105] 0 25 4 0 2 15 4 104 5 6 ...
## .. ..- attr(*, "dimnames")=List of 2
## ..$ replication_2: num [1:105, 1:105] 0 23 4 0 2 15 4 104 5 6 ...
## .. ..- attr(*, "dimnames")=List of 2
## ..$ replication_3: num [1:105, 1:105] 0 23 4 0 2 15 4 103 5 6 ...
## .. ..- attr(*, "dimnames")=List of 2
## ..$ proba : num [1:105, 1:105] 0.00 4.31e-05 2.16e-05 3.90e-06 1.81e-05 ...
## .. ..- attr(*, "dimnames")=List of 2
## $ parameter_2:List of 4
## ..$ replication_1: num [1:105, 1:105] 0 54 2 0 0 38 1 77 5 7 ...
## .. ..- attr(*, "dimnames")=List of 2
## ..$ replication_2: num [1:105, 1:105] 0 53 2 0 0 40 1 78 5 7 ...
## .. ..- attr(*, "dimnames")=List of 2
## ..$ replication_3: num [1:105, 1:105] 0 53 2 0 0 38 1 77 5 8 ...
## .. ..- attr(*, "dimnames")=List of 2
## ..$ proba : num [1:105, 1:105] 0.00 8.13e-05 8.17e-06 7.10e-07 3.16e-06 ...
## .. ..- attr(*, "dimnames")=List of 2
## - attr(*, "class")= chr [1:2] "TDLM" "list"
## - attr(*, "from")= chr "run_law_model"
## - attr(*, "proba")= logi TRUEIt is also important to note that the radiation law and the uniform law are free of parameter.
res <- run_law_model(law = "Rad",
mass_origin = mi,
mass_destination = mj,
distance = NULL,
opportunity = sij,
param = NULL,
write_proba = TRUE,
model = "DCM",
nb_trips = NULL,
out_trips = Oi,
in_trips = Dj,
average = FALSE,
nbrep = 3)
print(res)## Argument Value
## 1 Law Rad
## 2 Model DCM
## 3 #Replications 3The argument average can be used to generate an average
matrix based on a multinomial distribution (based on an infinite number
of drawings). In this case, the models’ inputs can be either positive
integer or real numbers and the output (nbrep = 1 in this
case) will be a matrix of positive real numbers.
res$replication_1[1:10,1:10]## [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
## [1,] 0 34 0 0 0 120 0 1 0 0
## [2,] 576 0 0 0 0 22 0 0 0 0
## [3,] 0 0 0 0 0 0 197 0 0 0
## [4,] 0 0 0 0 0 0 0 0 0 0
## [5,] 0 0 0 1 0 0 0 3 0 0
## [6,] 17 2 0 0 0 0 0 0 0 0
## [7,] 0 0 201 0 0 0 0 0 0 0
## [8,] 4 1 1 1 10 5 1 0 0 1
## [9,] 0 0 0 0 0 0 0 40 0 0
## [10,] 0 0 0 0 0 0 0 13 0 0
res <- run_law_model(law = "Rad",
mass_origin = mi,
mass_destination = mj,
distance = NULL,
opportunity = sij,
param = NULL,
write_proba = TRUE,
model = "DCM",
nb_trips = NULL,
out_trips = Oi,
in_trips = Dj,
average = TRUE,
nbrep = 3)
print(res)## Argument Value
## 1 Law Rad
## 2 Model DCM
## 3 #Replications 1 (average)
res$replication_1[1:10,1:10]## [,1] [,2] [,3] [,4] [,5]
## [1,] 4.207454e-04 3.248924e+01 1.629984e-01 0.0169209490 1.523725e-01
## [2,] 5.645506e+02 6.155812e-04 1.486180e-01 0.0133929974 1.262074e-01
## [3,] 8.174148e-02 2.927128e-02 8.564946e-04 0.0180562693 1.785403e-01
## [4,] 5.866220e-03 1.762770e-03 5.461852e-03 0.0006206272 1.984250e-01
## [5,] 7.824940e-02 2.902253e-02 2.083665e-01 1.3841229993 7.860351e-04
## [6,] 1.794519e+01 2.896778e+00 1.227317e-01 0.0118216041 1.064531e-01
## [7,] 4.854529e-02 1.825077e-02 2.003666e+02 0.0121155517 1.240133e-01
## [8,] 4.235287e+00 1.150834e+00 1.817298e+00 1.5952317911 1.087001e+01
## [9,] 1.701860e-02 6.442397e-03 1.358021e-02 0.0027705322 2.718136e-02
## [10,] 3.657437e-01 9.929259e-03 1.029943e-02 0.0078265447 2.624542e-02
## [,6] [,7] [,8] [,9] [,10]
## [1,] 1.206594e+02 8.830093e-02 1.637304888 0.0120698542 0.0777923728
## [2,] 2.261365e+01 1.297312e-01 0.862290746 0.0050699294 0.0060667099
## [3,] 1.934135e-01 1.973623e+02 0.918724724 0.0061614808 0.0053164987
## [4,] 9.469349e-03 6.169547e-03 0.278155220 0.0008433002 0.0012892865
## [5,] 1.033249e-01 2.460352e-01 3.462295645 0.0158169340 0.0150487684
## [6,] 5.850044e-04 1.180099e-01 0.752499705 0.0038974570 0.0092130934
## [7,] 1.138611e-01 2.549294e-03 0.633540548 0.0040869895 0.0021031775
## [8,] 5.040588e+00 1.982879e+00 0.002650424 0.7993806174 1.0370497229
## [9,] 2.296625e-02 1.230871e-02 39.049317001 0.0005691793 0.0015470504
## [10,] 6.732294e-02 1.102089e-02 13.987963900 0.0025478503 0.0002716179The functions run_law and run_model have been designed to run only one of the two components of the two-step approach. They function the same as a run_law_model, but it is worth noting that only inter-location flows are considered for the distribution laws, meaning that the matrix of probabilities (and associated simulated OD matrices) generated by a given distribution law with run_law_model or run_law is a zero-diagonal matrix. Nevertheless, it is possible to generate intra-location flows with run_model taking any kind of matrix of probabilities as input.
Parameters’ calibration & models’ evaluation
The package contains two function to help calibrating and evaluating the model. The function gof computes goodness-of-fit measures between observed and simulated OD matrices and the function calib_param that estimates the optimal parameter value for a given law and a given spatial distribution of location based on the Figure 8 in (Lenormand et al., 2016).
Let us illustrate the trip distribution laws and models’ calibration with the the normalized gravity law with an exponential distance decay function and the Doubly Constrained Model. Based on the average surface area of the Kansas counties (in square kilometers) it seems that the optimal parameter value of the normalized gravity law with an exponential distance decay function (as described in (Lenormand et al., 2016)) for commuting in US Kansas counties is around 0.085.
print(calib_param(av_surf = mean(spi$surface), law = "NGravExp"))## [1] 0.08521097This is just an estimation that help us to identify the potential range of parameter value, so in order to rigorously calibrate and evaluate the trip distribution law and model we need to compute the goodness-of-fit measure for a wide range of parameter values.
res <- run_law_model(law = "NGravExp",
mass_origin = mi,
mass_destination = mj,
distance = distance,
opportunity = NULL,
param = seq(0.05,0.1,0.005),
write_proba = TRUE,
model = "DCM",
nb_trips = NULL,
out_trips = Oi,
in_trips = Dj,
average = FALSE,
nbrep = 3)
calib <- gof(sim = res, obs = od, measures = "all", distance = distance)
print(calib)## Parameter Parameter_value Simulation CPC NRMSE KL
## 1 parameter_1 0.05 replication_1 0.8163786 10.506334 0.12959589
## 2 parameter_1 0.05 replication_2 0.8165034 10.510550 0.12948446
## 3 parameter_1 0.05 replication_3 0.8163337 10.498985 0.12967025
## 4 parameter_2 0.055 replication_1 0.8317619 9.400138 0.11352553
## 5 parameter_2 0.055 replication_2 0.8316072 9.419131 0.11319080
## 6 parameter_2 0.055 replication_3 0.8316122 9.421604 0.11328522
## 7 parameter_3 0.06 replication_1 0.8438958 8.652907 0.09821787
## 8 parameter_3 0.06 replication_2 0.8441254 8.631445 0.09800527
## 9 parameter_3 0.06 replication_3 0.8441304 8.625002 0.09796417
## 10 parameter_4 0.065 replication_1 0.8519668 8.171437 0.08799202
## 11 parameter_4 0.065 replication_2 0.8520816 8.164006 0.08777108
## 12 parameter_4 0.065 replication_3 0.8521415 8.174424 0.08806166
## 13 parameter_5 0.07 replication_1 0.8551912 7.960820 0.08028067
## 14 parameter_5 0.07 replication_2 0.8553060 7.964459 0.08026958
## 15 parameter_5 0.07 replication_3 0.8553310 7.957509 0.08016854
## 16 parameter_6 0.075 replication_1 0.8556704 7.952719 0.07954942
## 17 parameter_6 0.075 replication_2 0.8555806 7.962511 0.07928927
## 18 parameter_6 0.075 replication_3 0.8557253 7.954575 0.07916219
## 19 parameter_7 0.08 replication_1 0.8549966 8.120059 0.07456592
## 20 parameter_7 0.08 replication_2 0.8551313 8.102435 0.07430036
## 21 parameter_7 0.08 replication_3 0.8551064 8.117285 0.07433799
## 22 parameter_8 0.085 replication_1 0.8529751 8.392804 0.07382928
## 23 parameter_8 0.085 replication_2 0.8530899 8.371318 0.07372981
## 24 parameter_8 0.085 replication_3 0.8530949 8.378601 0.07370104
## 25 parameter_9 0.09 replication_1 0.8498056 8.716578 0.07529957
## 26 parameter_9 0.09 replication_2 0.8499553 8.706906 0.07521192
## 27 parameter_9 0.09 replication_3 0.8498705 8.716180 0.07526508
## 28 parameter_10 0.095 replication_1 0.8463815 9.090134 0.08095543
## 29 parameter_10 0.095 replication_2 0.8463017 9.087384 0.08112540
## 30 parameter_10 0.095 replication_3 0.8462617 9.104245 0.08110074
## 31 parameter_11 0.1 replication_1 0.8424134 9.474920 0.08345803
## 32 parameter_11 0.1 replication_2 0.8423186 9.490023 0.08326501
## 33 parameter_11 0.1 replication_3 0.8423485 9.483437 0.08345601
## CPL CPC_d KS_stat KS_pval
## 1 0.6590965 0.8954314 0.07394171 0.9998142
## 2 0.6592593 0.8953166 0.07407648 0.9998070
## 3 0.6595955 0.8953166 0.07412639 0.9998047
## 4 0.6716883 0.9156713 0.05089170 1.0000000
## 5 0.6715215 0.9155914 0.05091666 1.0000000
## 6 0.6713432 0.9155615 0.05099153 1.0000000
## 7 0.6718877 0.9316536 0.03124579 1.0000000
## 8 0.6722507 0.9318083 0.03122582 1.0000000
## 9 0.6720692 0.9317235 0.03125078 1.0000000
## 10 0.6726710 0.9430788 0.03475470 1.0000000
## 11 0.6724866 0.9431337 0.03475470 1.0000000
## 12 0.6728604 0.9430837 0.03474971 1.0000000
## 13 0.6744594 0.9493878 0.03844829 1.0000000
## 14 0.6744526 0.9495725 0.03845827 1.0000000
## 15 0.6742623 0.9495725 0.03835346 1.0000000
## 16 0.6720867 0.9479803 0.04456268 1.0000000
## 17 0.6716867 0.9481200 0.04458764 1.0000000
## 18 0.6718891 0.9480801 0.04459762 1.0000000
## 19 0.6633633 0.9408776 0.05041253 1.0000000
## 20 0.6631546 0.9409175 0.05048740 1.0000000
## 21 0.6623336 0.9407079 0.05053233 1.0000000
## 22 0.6516067 0.9328265 0.05747528 1.0000000
## 23 0.6513849 0.9329913 0.05739542 1.0000000
## 24 0.6513849 0.9329563 0.05744034 1.0000000
## 25 0.6406504 0.9257189 0.06334510 0.9999989
## 26 0.6406504 0.9258487 0.06332513 0.9999989
## 27 0.6408588 0.9257538 0.06337005 0.9999988
## 28 0.6355202 0.9187060 0.06824160 0.9999913
## 29 0.6353097 0.9186911 0.06822663 0.9999914
## 30 0.6350679 0.9186262 0.06833644 0.9999910
## 31 0.6240271 0.9121624 0.07363724 0.9999478
## 32 0.6237724 0.9120476 0.07377201 0.9999454
## 33 0.6237724 0.9120626 0.07376202 0.9999458All the necessary information is stored in the object calib, most of the goodness-of-fit measures agree on a parameter value of 0.075 in that case with an associated average Common Part of Commuter equal to 85.6%.
cpc <- aggregate(calib$CPC, list(calib$Parameter_value), mean)[,2]
oldmar <- par()$mar
par(mar = c(4.5, 6, 1, 1))
plot(seq(0.05,0.1,0.005), cpc, type="b", pch=16, cex=2, lty=1, lwd=3,
col="steelblue3", axes=FALSE, xlab="", ylab="")
axis(1, cex.axis=1.2)
axis(2, cex.axis=1.2, las=1)
mtext("Parameter value", 1, line = 3.25, cex = 1.75)
mtext("Common Part of Commuters", 2, line = 4, cex = 1.75)
box(lwd=1.5)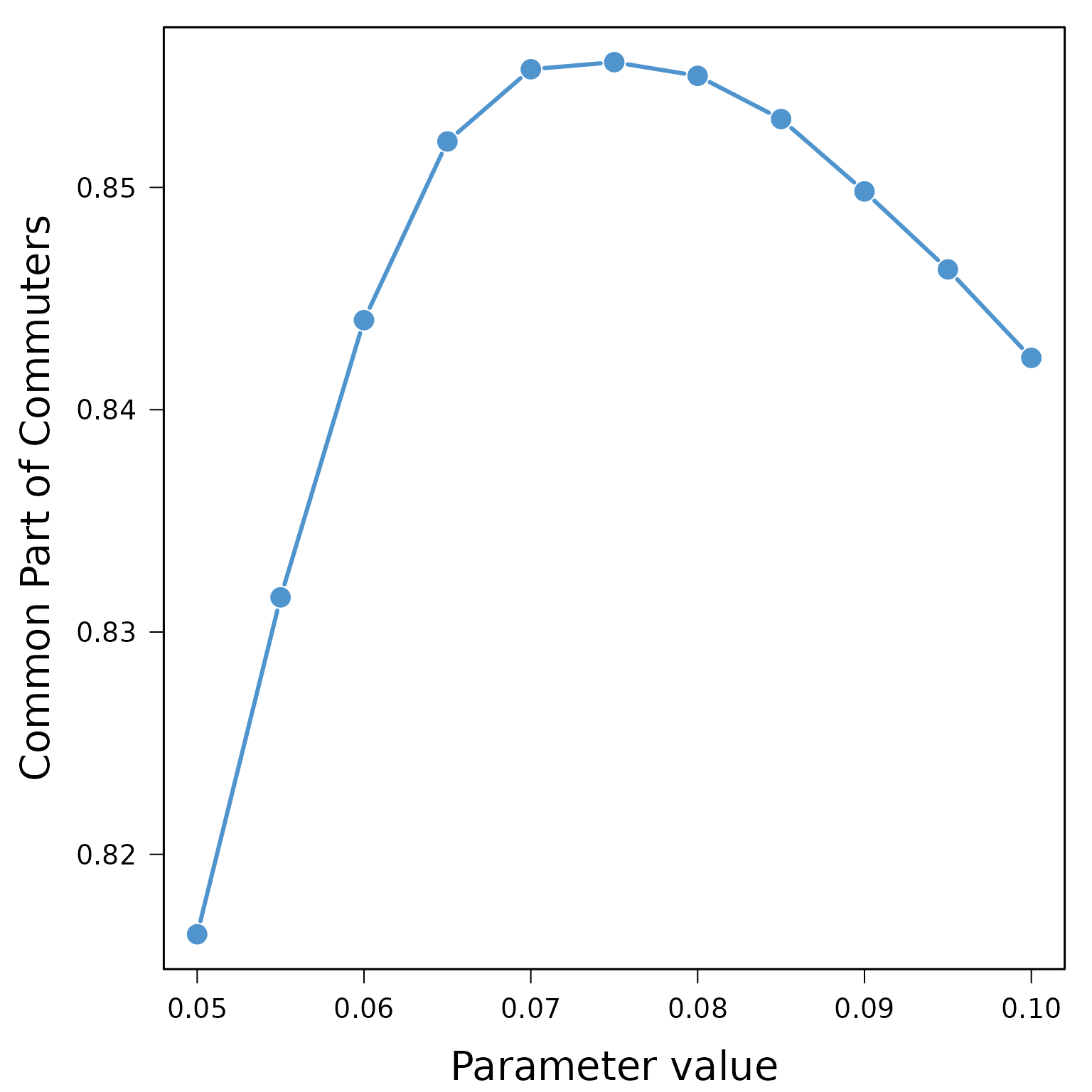
par(mar = oldmar)